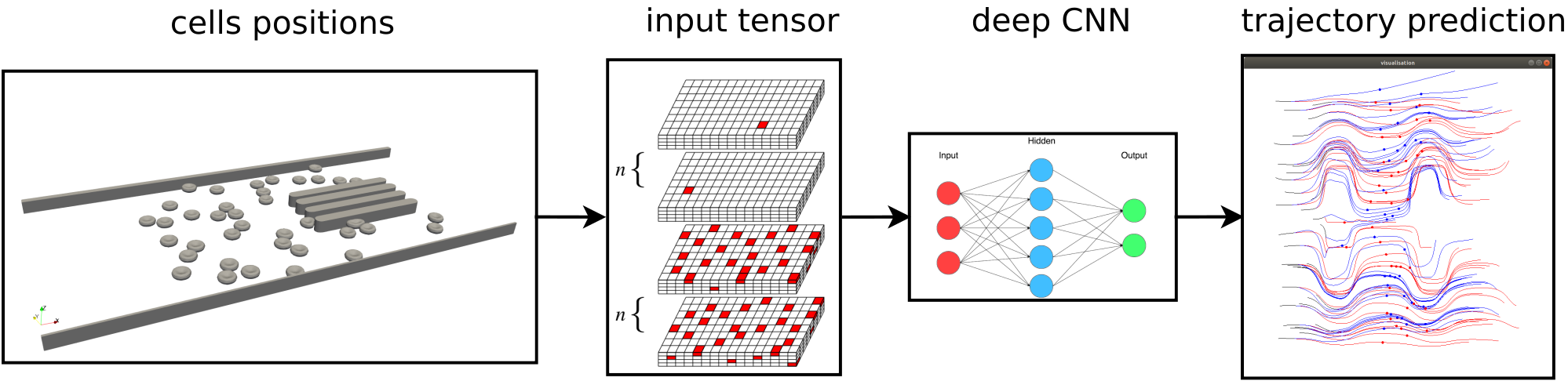
 Using a computational model to examine blood flow at cell-scale, including the elasticity of the red blood cells, can be done quite precisely in small channels and within a short timeframe. However, larger volumes or timescales require different approaches. Instead of simplifying the simulation, we use a neural network to predict the movement of the red blood cells.
Using a computational model to examine blood flow at cell-scale, including the elasticity of the red blood cells, can be done quite precisely in small channels and within a short timeframe. However, larger volumes or timescales require different approaches. Instead of simplifying the simulation, we use a neural network to predict the movement of the red blood cells.
The neural network still uses data from the numerical simulation for learning, however, the simulation needs only be run once. Alternatively, the data could come from video processing of a recording of a biological experiment. Afterwards, the network is able to predict the movement of the red blood cells. While for uncomplicated box channels there is no advantage in using neural networks instead of a simple prediction using fluid streamlines, for a more complicated geometry, the neural network is significantly more accurate. Another application is using it as a comparison tool for different modelled situations.
First, we used a framework inspired by the radial basis function network and Kohonen’s self-organized maps [1]. In [2], we advanced to deep learning model using convolutional neural networks. We evaluated several types of neural networks architectures, formats of their input data and the learning methods on simulation data inspired by a laboratory experiment.
More info:
- [1] H. Bachraty, K. Bachrata, M. Chovanec, I. Jancigova, M. Smieskova and K. Kovalcikova: Applications of machine learning for simulations of red blood cells in microfluidic devices, 2018, to appear in BMC Systems Biology
- [2] Chovanec M., Bachratý H., Jasenčáková K., Bachratá K. (2019) Convolutional Neural Networks for Red Blood Cell Trajectory Prediction in Simulation of Blood Flow. In: Rojas I., Valenzuela O., Rojas F., Ortuño F. (eds) Bioinformatics and Biomedical Engineering. IWBBIO 2019. Lecture Notes in Computer Science, vol 11466. Springer, Cham