 |
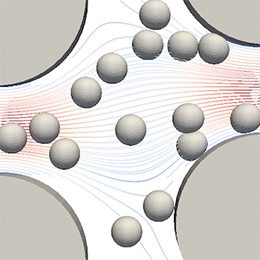
Computational model and its propertiesWe model elastic objects, mostly cells, in fluid flow in solid channels (microfluidic devices). We have developed Object-in-fluid framework as part of extensible open source package ESPResSo (www.espressomd.org). The dynamics of the fluid is modeled by the lattice-Boltzmann method (LBM). The LBM is fast and easily parallelizable, which is necessary for simulations that contain large number of immersed objects. Model of elastic object is based on a triangular spring mesh on the object’s surface. In our model we consider the mesh points to be connected with springs that exert forces according to their prolongation. We also investigate cell membrane viscosity and viscosity inside the cell. |
 |
Applications of machine learning for simulationsSimulations with many cells are very computationally intensive. We consider neural network predictions of cell trajectories and characteristics based on previous simulations. |
 |
Image processing of biological images and videosTo compare the simulation results with biological experiments, we develop methods for extracting cell information from images and videos of biological experiments – cell flows in microfluidic devices. |
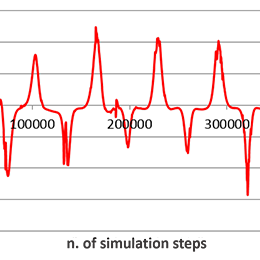 |
Statistical postprocessing of simulation dataStatistical processing of simulation results is done in order to understand the behavior of multi-cellular simulations. One-cell simulation can be compared with the corresponding biological experiment directly, by comparing individual dimensions or other attributes of the simulated and biological cell. In case of multi-cellular simulations, we need to use statistical tools to understand whether the numerical simulation has a similar behavior as the biological experiment. |
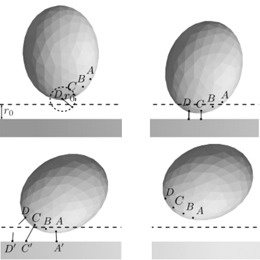 |
Affinity and adhesionThe biological concepts of adhesion rely on creating receptor-ligand (R-L) complexes between the receptor patches located on the surface of the cell and ligands distributed on the functionalized surfaces. |
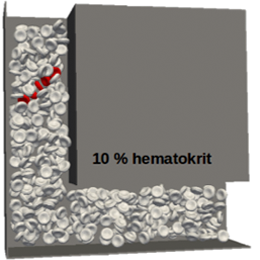 |
Applications of the computational modelWe have used the model to investigate capture rates of rare cells in periodic obstacle arrays, collision rates in shifted arrays, cell damage index of cells passing through different bends in channels, … |
Current research grants
Past research grants
|
|